Reliable medical image classification requires accurate predictions and well-calibrated uncertainty estimates, especially in high-stakes clinical settings. This work presents MedSymmFlow, a generative-discriminative hybrid model built on Symmetrical Flow Matching, designed to unify classification, generation, and uncertainty quantification in medical imaging.
MedSymmFlow leverages a latent-space formulation that scales to high-resolution inputs and introduces a semantic mask conditioning mechanism to enhance diagnostic relevance. Unlike standard discriminative models, it naturally estimates uncertainty through its generative sampling process. The model is evaluated on four MedMNIST datasets, covering a range of modalities and pathologies. The results show that MedSymmFlow matches or exceeds the performance of established baselines in classification accuracy and AUC, while also delivering reliable uncertainty estimates validated by performance improvements under selective prediction.
MedSymmFlow introduces a symmetrical flow-matching framework for joint semantic image synthesis and classification. It models bidirectional transformations between an image X and its semantic representation Y by learning a joint velocity field that drives both forward (from semantic input to image) and reverse (from image to semantic prediction) flows. This allows the model to treat both generation and prediction as a continuous integration process over time, unifying the tasks within a single dynamic system.
Instead of using one-hot or grayscale class labels, MedSymmFlow adopts an RGB encoding scheme, where each class is assigned a unique RGB triplet. During training, these RGB labels are perturbed with small uniform noise to make the conditioning space continuous, allowing for more expressive flow matching. The model learns to transport these RGB-coded semantic targets across time alongside the image.
At inference time, the predicted semantic output lies in a continuous RGB space. To assign a class label, the model compares the output against the set of predefined RGB codes using Euclidean distance and selects the closest match. This process not only yields a hard classification label but also provides a measure of uncertainty based on the distance from the predicted color to the nearest class prototype.
To handle high-resolution medical images (up to 224×224 pixels), MedSymmFlow operates in a compressed latent space. It leverages the encoder and decoder from the Stable Diffusion Variational Autoencoder (VAE), reducing the dimensionality of both the input image and its RGB mask. These latent representations are then processed by the model, enabling efficient training and inference while preserving semantic fidelity.
After semantic prediction in latent space, the result is decoded back into the RGB domain using the VAE decoder. This design allows the model to scale to complex medical datasets while maintaining high generation quality and segmentation accuracy.
Table 1 presents classification results across four MedMNIST datasets. At a resolution of 28×28 pixels, the original SymmFlow framework—using grayscale masks—performs reasonably well on PneumoniaMNIST and BloodMNIST, but drops significantly in performance on DermaMNIST and RetinaMNIST. This is particularly evident in the AUC scores, and stems from a core limitation: grayscale encoding compresses all semantic class information along a single axis, limiting the model’s ability to capture complex multi-class relationships.
| Model | PneumMNIST | BloodMNIST | DermaMNIST | RetinaMNIST | ||||
|---|---|---|---|---|---|---|---|---|
| AUC | ACC | AUC | ACC | AUC | ACC | AUC | ACC | |
| ResNet-18 (28) | 94.4 | 85.4 | 99.8 | 95.8 | 91.7 | 73.5 | 71.7 | 52.4 |
| ResNet-50 (28) | 94.8 | 85.4 | 99.7 | 95.6 | 91.3 | 73.5 | 72.6 | 52.8 |
| auto-sklearn (28) | 94.2 | 85.5 | 98.4 | 87.8 | 90.2 | 71.9 | 69.0 | 51.5 |
| AutoKeras (28) | 94.7 | 87.8 | 99.8 | 96.1 | 91.5 | 74.9 | 71.9 | 50.3 |
| ResNet-18 (224) | 95.6 | 86.4 | 99.8 | 96.3 | 92.0 | 75.4 | 71.0 | 49.3 |
| ResNet-50 (224) | 96.2 | 88.4 | 99.7 | 95.0 | 91.2 | 73.1 | 71.6 | 51.1 |
| MedViT-S (224) | 99.5 | 96.1 | 99.7 | 95.1 | 93.7 | 78.0 | 77.3 | 56.1 |
| SymmFlow (28) | 91.6 ± 0.8 | 89.4 ± 0.5 | 99.1 ± 0.1 | 96.3 ± 0.2 | 83.4 ± 0.6 | 69.3 ± 0.5 | 70.2 ± 1.1 | 50.7 ± 0.7 |
| MSF (Ours) (28) | 95.2 ± 0.4 | 88.0 ± 0.6 | 99.4 ± 0.1 | 97.9 ± 0.2 | 89.6 ± 0.5 | 78.8 ± 0.8 | 73.1 ± 0.4 | 51.4 ± 2.1 |
| LatMSF (Ours) (224) | 94.4 ± 1.7 | 89.4 ± 1.1 | 99.8 ± 0.0 | 99.0 ± 0.1 | 92.5 ± 0.4 | 81.0 ± 0.6 | 78.8 ± 0.4 | 54.0 ± 0.9 |
RGB mask conditioning in MedSymmFlow directly resolves this limitation. By mapping class information into structured RGB vectors, the model can distinguish categories more effectively, which leads to clear gains in both AUC and classification accuracy—especially in datasets like DermaMNIST where fine-grained class separability is critical.
Extending this approach to latent space via LatMSF amplifies these benefits. The model consistently matches or outperforms ViT-based classifiers, while outperforming all CNN and AutoML baselines across multiple tasks. The one exception is PneumoniaMNIST, where performance drops—likely due to the fixed VAE encoder failing to preserve key discriminative cues.
While LatMSF achieves top-tier results, this comes at the cost of increased inference time and memory usage, driven primarily by VAE sampling and decoding overhead. Addressing this trade-off will be a focus of future optimization efforts.
Figure below illustrates that for both the low- and high-resolution variants, confidence correlates with accuracy—indicating that the proposed distance-based uncertainty proxy is well calibrated and can be used to reject low-quality predictions. When the most uncertain predictions are removed, classification accuracy improves consistently across datasets, demonstrating that the model is capable of accurately assessing its own prediction reliability.
This behavior is particularly useful in high-stakes settings such as medical imaging, where abstaining from uncertain predictions is preferable to making incorrect confident ones. The effect is most pronounced in PneumoniaMNIST and RetinaMNIST. However, overfiltering begins to harm performance in DermaMNIST at higher resolution, as some confident and correct predictions are also discarded. BloodMNIST shows minimal impact, as accuracy is already high without uncertainty-based filtering.
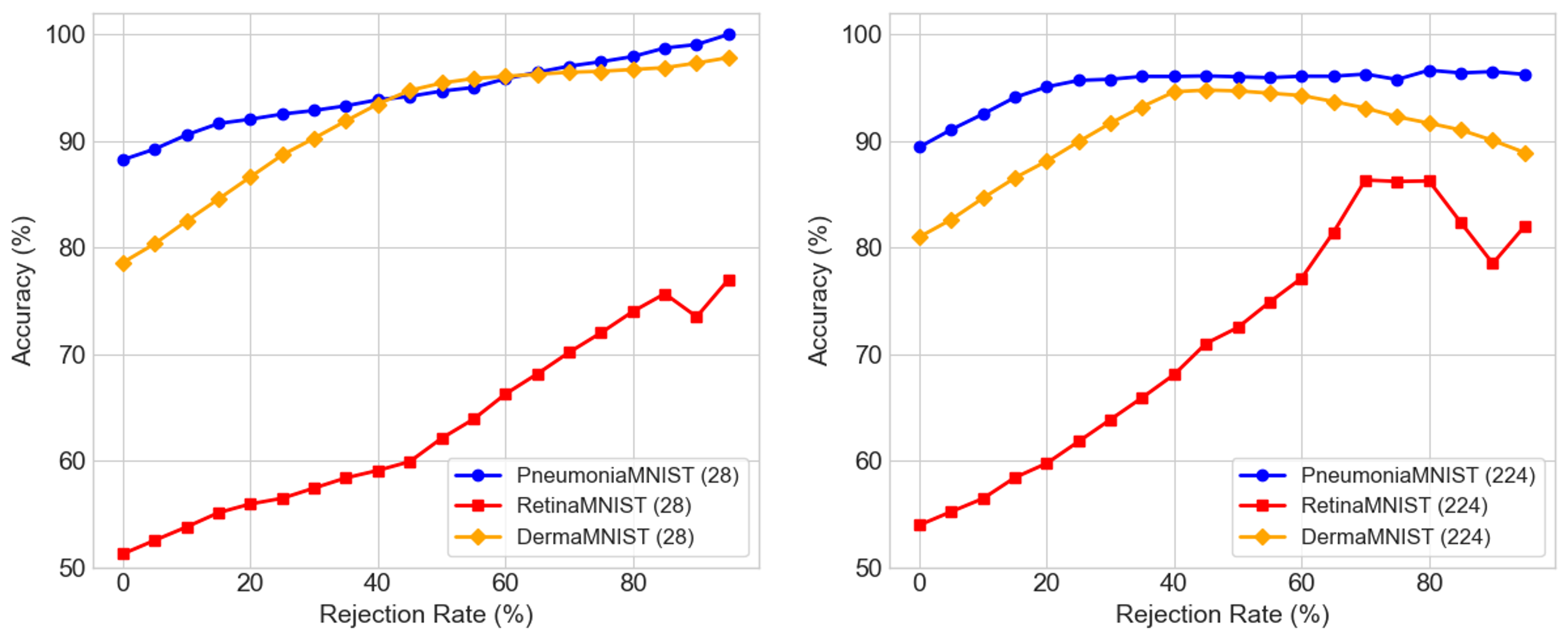
Filtering uncertain predictions improves accuracy, confirming that the distance-based uncertainty measure is well calibrated.
The figure below presents high-resolution samples generated using the latent-space variant of MedSymmFlow. These outputs demonstrate strong visual fidelity across multiple datasets. Notably, the model accurately captures fine-grained semantic features such as skin texture and hair artifacts in DermaMNIST, as well as retinal vessel structure and lesion contrast in RetinaMNIST. This indicates that the model not only classifies effectively, but also learns a semantically grounded, dataset-specific representation capable of producing realistic, diverse samples that reflect both anatomical structure and pathological variation.
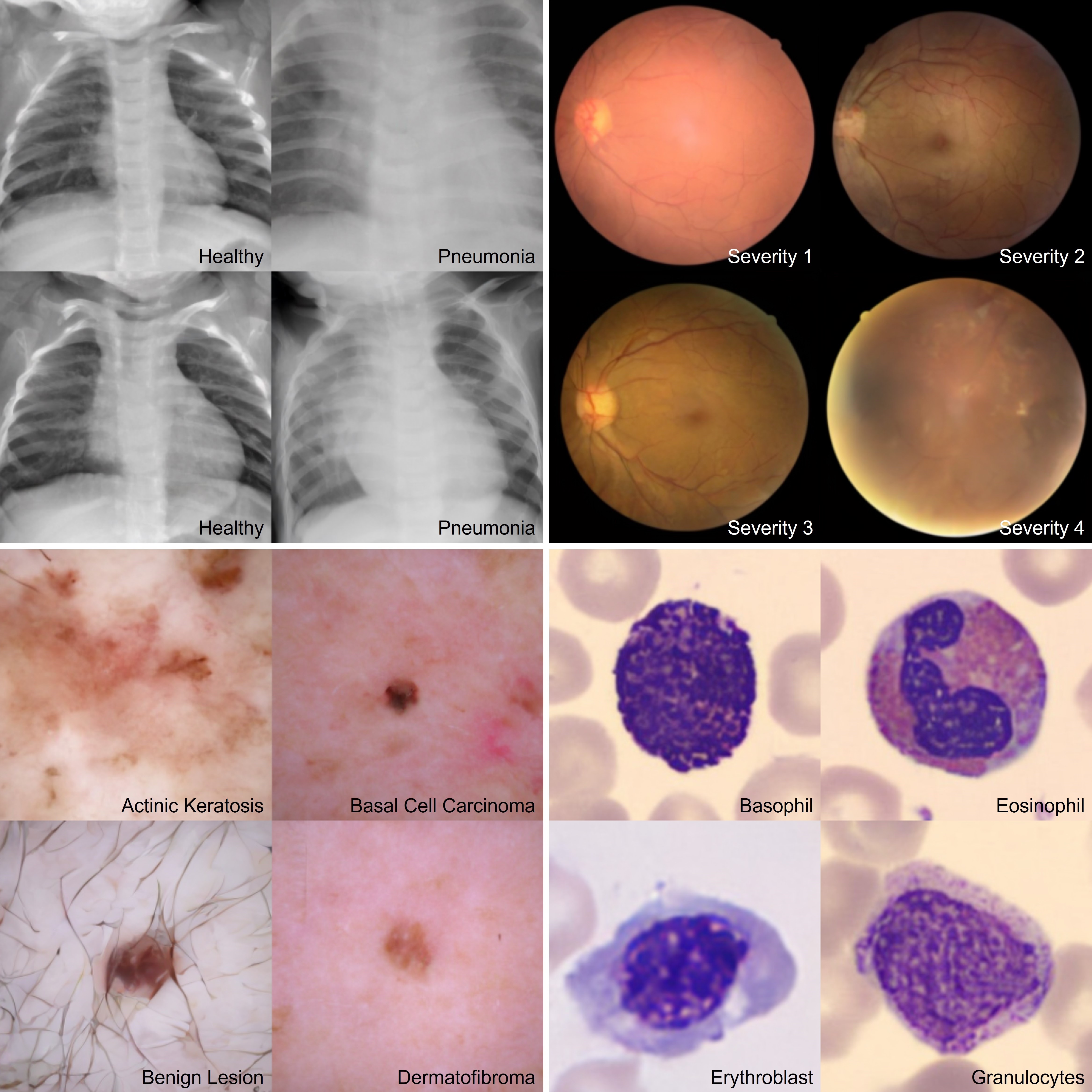
Samples generated by the latent models trained on the four selected MedMNIST datasets, using an Euler ODE solver with 25 steps.
@inproceedings{caetano2025medsymmflow,
title={MedSymmFlow: Bridging Generative Modeling and Classification in Medical Imaging through Symmetrical Flow Matching},
author={Caetano, Francisco and Abdi, Lemar and Viviers, Christiaan and Valiuddin, Amaan and van der Sommen, Fons},
booktitle={MICCAI Workshop on Deep Generative Models},
pages={35--45},
year={2025},
organization={Springer}
}